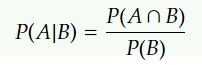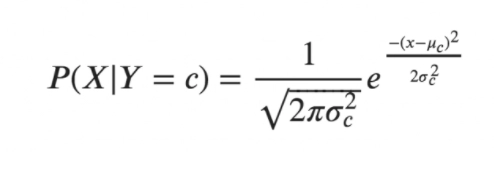Gaussian Naive Bayes
We've seen that the events(A1, A2,...) are in categories so far, but what about when an event is a continuous variable? If we suppose that event follows a specific distribution, the probability of likelihoods are calculated using the probability density function of that distribution.
sourceLink
If we assume that events follow a Gaussian or normal distribution, we must use its probability density and call it Gaussian Naive Bayes.
For the Naive gaussian Bayes, we use the following form:
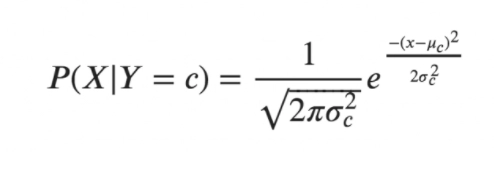
The variance and mean of the continuous variable X determined for a given class c of Y are σ and μ in the following formulas.
Let's use Python,, and Scikit Learn to implement Gaussian Naive Bayes in practice.
Implementation
%matplotlib inline
From sklearn.datasets import make_blobs, make_moons, make_regression, load_iris
from scipy.stats import multivariate_normal
import matplotlib.pyplot as plt
import numpy as np
# For the moment we only take a couple of features from the IRIS dataset for convenience of visualization
iris = load_iris()
X = iris.data[:, 1:3]
Y = iris.target
print X.shape
print Y.shape
plt.scatter(X[:,0],X[:,1])
#algorithm implementation.
class BayesClassifier:
mu = None
cov = None
n_classes = None
def __init__(self):
a = None
def pred(self,x):
prob_vect = np.zeros(self.n_classes)
for i in range(self.n_classes):
mnormal = multivariate_normal(mean=bc.mu[i], cov=bc.cov[i])
# We use uniform priors
prior = 1./self.n_classes
prob_vect[i] = prior*mnormal.pdf(x)
sumatory = 0.
for j in range(self.n_classes):
mnormal = multivariate_normal(mean=bc.mu[j], cov=bc.cov[j])
sumatory += prior*mnormal.pdf(x)
prob_vect[i] = prob_vect[i]/sumatory
return prob_vect
def fit(self, X,y):
self.mu = []
self.cov = []
self.n_classes = np.max(y)+1
for i in range(self.n_classes):
Xc = X[y==i]
mu_c = np.mean(Xc, axis=0)
self.mu.append(mu_c)
cov_c = np.zeros((X.shape[1], X.shape[1]))
for j in range( Xc.shape[0]):
a = Xc[j].reshape((X.shape[1],1))
b = Xc[j].reshape((1,X.shape[1]))
cov_ci = np.multiply(a, b)
cov_c = cov_c+cov_ci
cov_c = cov_c/float(X.shape[0])
self.cov.append(cov_c)
self.mu = np.asarray(self.mu)
self.cov = np.asarray(self.cov)
# Fit the classifier
bc = BayesClassifier()
bc.fit(X,Y)

You can also try this code with Online Python Compiler
FAQs
-
Is Gaussian naive Bayes suitable for datasets with a large number of attributes??
If the number of characteristics is enormous, the computation cost will be considerable, and the Curse of Dimensionality will apply.
-
What is the zero frequency phenomenon?
The model is given a 0 (zero) probability if a category is not recorded in the training set but occurs in the test data set, resulting in an erroneous estimate.
-
What exactly does the word distribution signify?
Distribution shows how values disperse in series and how frequently they appear in this series.
-
When is gaussian naive bayes used?
It is more likely to be used when each class follows a Gaussian distribution.
-
What is a confusion matrix?
A confusion matrix is a method for evaluating machine learning classification performance. It allows you to assess the performance of the classification model using a set of test data for which the true and false values are known.
Key Takeaways
There are several different naive Bayes algorithms whose usability can be referred to here.