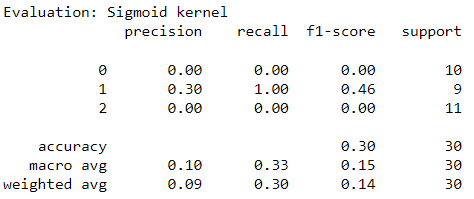
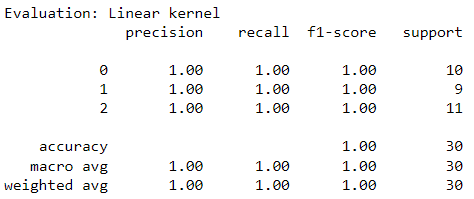
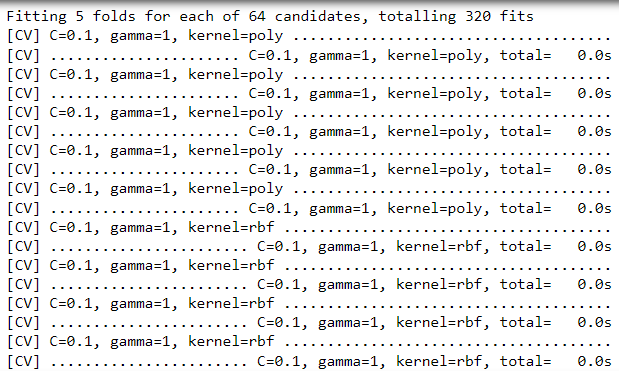
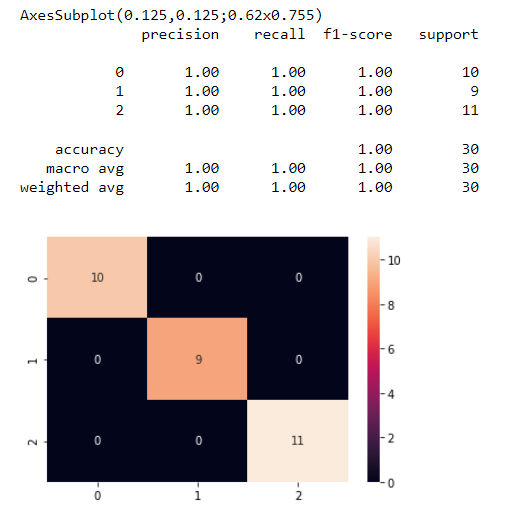
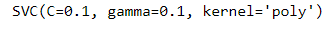Introduction
Do you want to know about the machine learning algorithm that can be used in both face detection and Bioinformatics? Well, you have come to the right place! In this article, we will talk about Support Vector Machines. Not only this, but we are also going to see how we can make our model highly accurate by changing its hyperparameters. However, finding the optimal values for these hyperparameters can be cumbersome and time-consuming if you do it manually. So, what should we do to make this task easier? We use GridSearchCV! Let’s discuss SVM, SVM hyperparameters, and their tuning with GridSearchCV both theoretically and practically!
Support Vector Machine and its basic intuition
Support vector machines fall under supervised machine learning techniques that are mainly used for classification problems. You can use it for regression problems too.
The basic idea behind the Support vector machine is that it takes data of relatively low dimension and converts it into higher dimensional data. Then it classifies the data into different categories by separating the data using a Hyperplane.
Let us understand the basic intuition behind a support vector machine using an example:
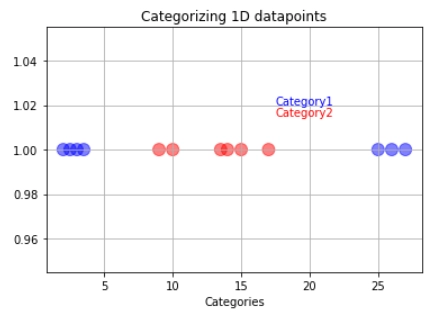
Let us imagine that we have a training dataset. The training data have been divided into category one (blue) and category two (red).
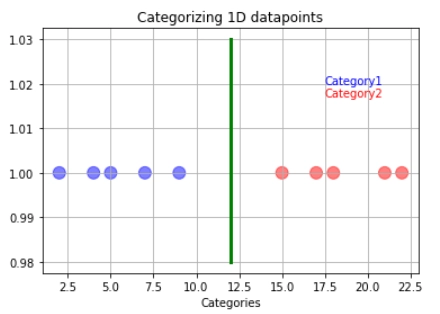
If we pass our training data through our model, it will set a decision boundary/threshold in the center of the last blue and the first red observations. The threshold is shown with the green line.
Note: The decision boundary, in this case, is a point, but it has been represented with a line to be visible properly without leading to confusion.
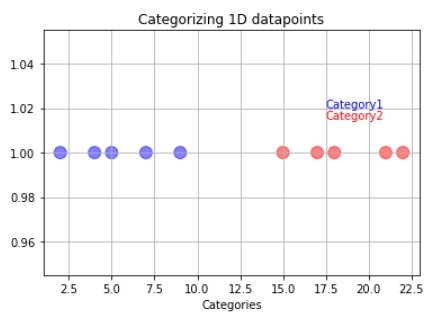
So if new data comes, we can easily classify it into either category one or category two depending on which side of the line they are.
But what happens if the training data has outliers? Suppose our model follows the hard and fast rule of making a boundary directly at the middle of the last category one and first category two observations.
In that case, this may lead to wrong predictions regarding the testing data. In this example, the orange data point represents the new data.
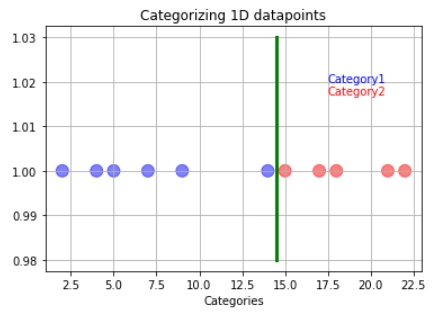
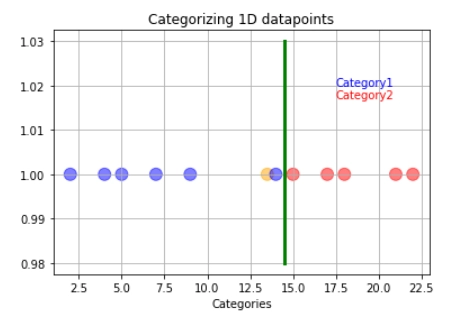
The orange data will get classified as blue instead of red even though it is closer to the red data point. So how do we overcome the problem of outliers? It is simple: We just have to allow misclassifications!
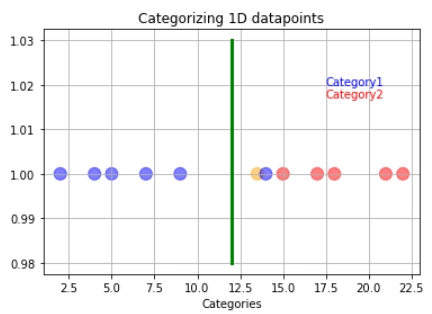
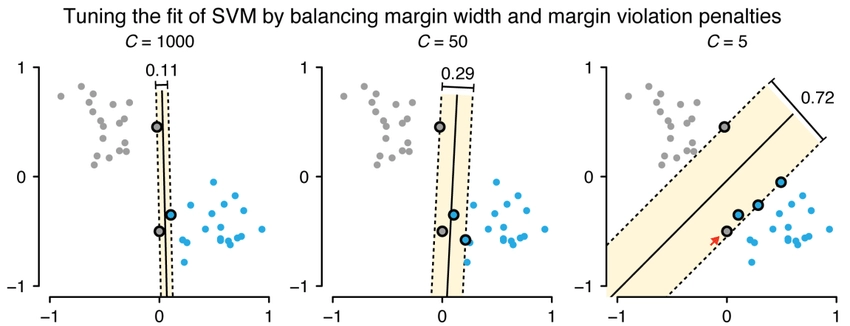
We allow the outlier to be misclassified as red data to have a better decision boundary. When we allow misclassification within the margin, the margin is called a soft margin.
In the case above, what we are using is a support vector classifier. We are using the support vector classifier to find the suitable decision boundary by allowing misclassifications.
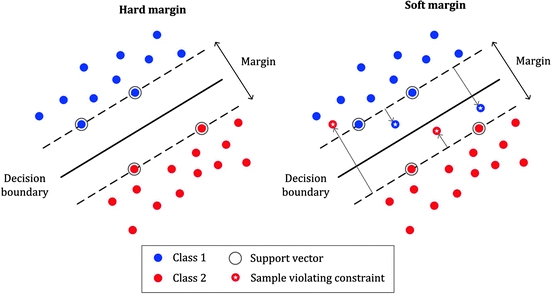
Allowing misclassification is a part bias-variance trade-off as we put some of the training data into the wrong category so that our model works well on testing data.
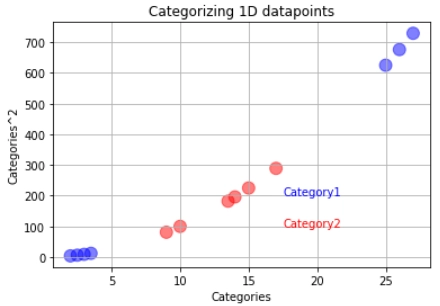
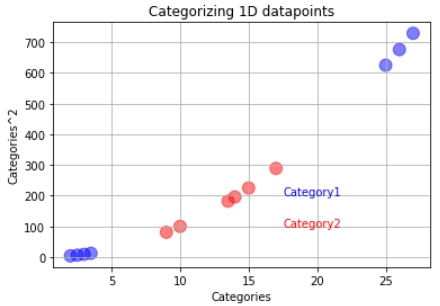
But what if our training data looks like below:
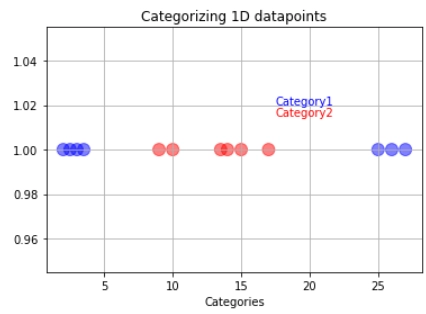
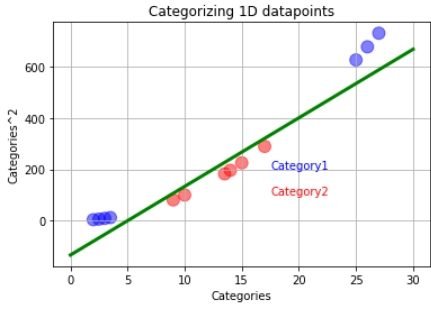
In a case like this, we will use the support vector machines. The support vector machine first converts the lower dimension training data into higher dimensional data. In this case, from one dimension, the data gets converted into two-dimension. How is this done? The square of the data becomes the y axis.
After this, we find the suitable decision boundary using the support vector classifier. In this case, our decision boundary becomes a two-dimensional hyperplane.
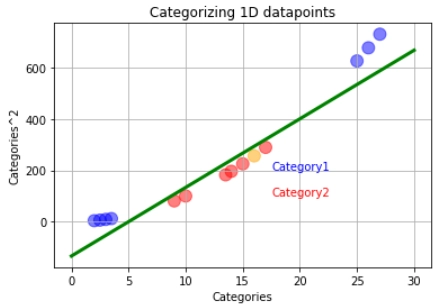
If new data comes, the support vector machine will square it and classify it depending on which side of the hyperplane it is. The orange data represents the latest data. It will be classified into the red category.
The code for all the graphs is given below:
#imports
import pandas as pd
import numpy as np
from sklearn import datasets
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.model_selection import GridSearchCV
import matplotlib.pyplot as plt
import seaborn as sns
#So what should we do to make our classifier not sensitive to outliers?
#WE SHOULD ALLOW MISSCLASSIFATION
x7=[2,4,5,7,9,13.5,14,15,17,18,21,22]
y7=np.ones(12)
size=np.ones(12)*120
colors=['b','b','b','b','b','orange','b','r','r','r','r','r']
plt.title("Categorizing 1D datapoints")
plt.xlabel("Categories")
plt.text(x=17.5, y=1.02, s="Category1",color='b')
plt.text(x=17.5, y=1.0170, s="Category2",color='r')
plt.scatter(x7,y7,s=size,c=colors,alpha=0.5)
#making the support vector classifier threshold with soft margin
x8=np.ones(4)*(((15-9)/2)+9)
y8=[0.98,1,1.02,1.03]
plt.plot(x8,y8,c='green',linewidth=3)
plt.grid()
print(plt.show())
#category1=[2,4,5,7,18,21,22]
#category2=[9,10,14,13.5,15,17]
#16 is the new data
x12=np.array([2,2.5,3,3.5,9,10,13.5,14,15,16,17,25,26,27])
y12=(x12)**2
size=np.ones(14)*120
colors=['b','b','b','b','r','r','r','r','r','orange','r','b','b','b']
plt.title("Categorizing 1D datapoints")
plt.xlabel("Categories")
plt.ylabel("Categories^2")
plt.text(x=17.5, y=200, s="Category1",color='b')
plt.text(x=17.5, y=100, s="Category2",color='r')
plt.scatter(x12,y12,s=size,c=colors,alpha=0.5)
#Finding the perfect support vector classifier
x13=np.array([0,5,10,15,20,25,30])
y13=((80/3)*x13)-(400/3) #calculated using simple line equation with two points
plt.plot(x13,y13,c='green',linewidth=3)
plt.grid()
print(plt.show())